The following sections contain the project specific details of the accepted proposal for the project: Patient Matching Module Strategy Enhancements.
Basic Understanding of the project
Following points briefly summarize my understanding of the project:
Patient Matching Module is a module which takes different data sources as input and identifies records which belong to the same patient. It is also used for the purpose of de-duplication in the same dataset.
The matching is done based on the fields of the dataset. Among the various fields in the dataset, which fields to use for matching is required as input in the Patient Matching Module as of now.
There are some statistics associated with each field (for example Hmax, UqVal etc) which are called field metrics. A domain expert can look at these field metrics and tell us which fields to use for Patient Matching.
We have a training dataset of these field metrics with the domain expert advice, based on that we want to build a forest of decision trees which we can use to check whether a field would be suitable for Patient Matching or not.
Proposed Approach
Following points summarize how I plan to approach the project:
- Given a dataset, calculate the field metrics (Primary mentor for this project, Jeremy, told me that the algorithms for these already exists, we would just need to implement those).
Some field metrics depend on the size of the dataset, for example Hmax, UqVal etc. Instead of considering their values, we would consider their percentage.
As I discussed with Jeremy, we have only one training dataset, instead of building the decision trees from the same dataset again and again, it would be better if we would just store the agreeable set of decision trees.
Jeremy has written a python code which builds the decision trees based on the training dataset. I would run that code and get the agreeable set of decision trees. After that I would encode the trees in a format we find best (probably xml). These decision trees would be resource to our system.
Having done that, I would write a code which would read the stored decision trees, take the field metrics (calculated from the dataset) as input and using the decision trees provide us the fields to use for Patient Matching.
- Then I will build a UI for this system and I will integrate the entire system with the Patient Matching Module.
Proposed Timeline
A rough project timeline is as follows:
Community Bonding Period (May 28 - June 16)
- Create a blog about the Patient Matching Module Strategy Enhancements project and send its URL to Michael Downey.
- Familiarize with OpenMRS module conventions.
- Go through the source code of Patient Matching Module. Figure out how to add and integrate this project with the module.
- Gather up the resources needed to start this project i.e. the algorithms for calculating the field metrics and Jeremy’s python code for building decision trees.
June 17 - June 23
Use Jeremy’s code to get an agreeable set of decision trees.
Encode the trees in a suitable format and save them for future use.
June 24 – July 5
Implement field metrics algorithms.
July 6 – August 5
Write a code which will take a dataset as input and output the fields suitable for patient matching. The code will use the encoded decision trees and the field metrics algorithms implemented before to do so.
I have classes starting from August 6th. They would be twelve hours a week. As suggested by Gaurav Paliwal, I have modified the project timeline to accommodate the time consumed by the classes.
August 6 – August 15
Unit testing of the system.
August 16 – August 31
Make a UI for the system.
September 1 – September 14
Integrate the system with Patient Matching Module.
September 15 – September 27
Integration Testing.
September 27 – October 10
Documentation.
Proposed UI
As a part of this GSoC project, a new module page “Suggested Strategy” (say) would be added to Patient Matching Module. The admin UI after this addition would look like:
When you navigate to this new module page “Suggested Strategy”, the page would look something like:
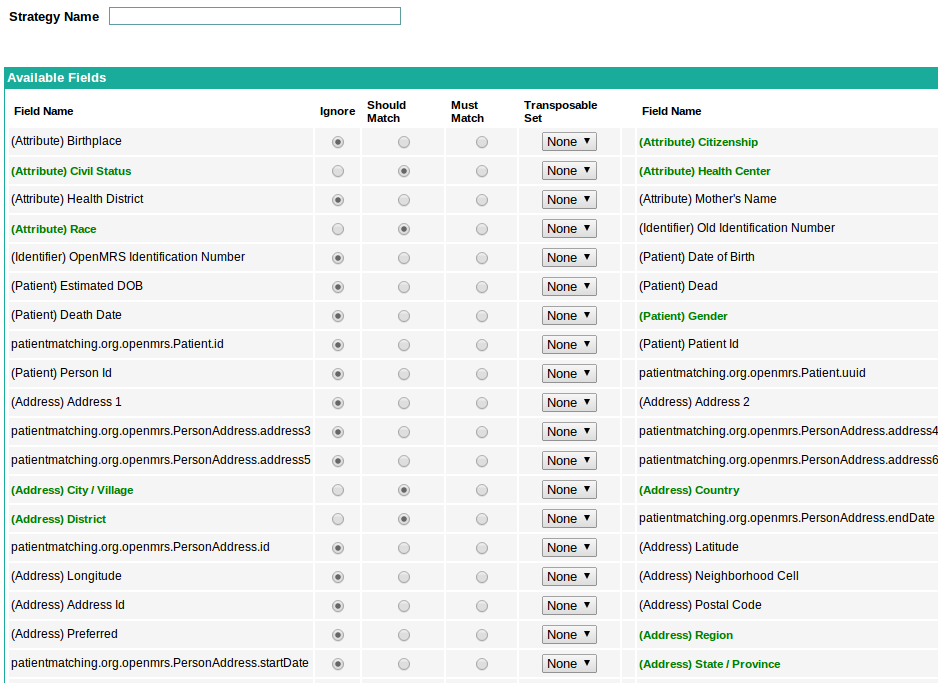
The page suggests attributes best suited for patient matching. I need to discuss with the mentors whether these suggested attributes are “Should Match” attributes or “Must Match” attributes or a mixture of both. Anyway, when the user clicks on “Get suggested attributes”, the resulting page contains all the attributes with the suggested attributes selected. The user can add to (and delete from) suggested attributes and then save the strategy. The resulting page would look something like:
The user has the facility to give the suggested strategy a strategy name, to modify the suggested strategy and to save the strategy.
Coding Assignment
Following was the assignment given by Gaurav Paliwal to me:
Write a small java program that read an XML file (a.XML) at a user defined location that location is inturn specified inside another XML file (b.XML) that is located in the same directory where your other java program files are.
Please host your code on github. Also commit code in github every hours. I want to see how you approach this assignment step by step, so commit early commit often.
My solution to the above assignment is hosted here: https://github.com/GarimaAhuja/ReadXML .