Introduction
Record linkage is the task of identifying pieces of scattered information that refer to the same thing. Patient matching is a specific application, in which we try to identify records that belong to the same patient among different data sources. These sources can range from patient data collected at different hospitals to external information from governmental institutions, such as death master file etc.
One of the interesting and challenging aspects of this project is to deal with erroneous data, for instance when your name is misspelled or your birth date is entered incorrectly. These kinds of things often happen in reality, and we can account for them by using flexible distance metrics and statistical models.
Why is then record linkage important and what are the benefits?
Well, we are living in an exciting period of globalization, where computers and internet make world-wide collaboration easy and necessary. Patient linkage and data aggregation techniques will allow medical institutions to store their own data, yet at the same time work together with others to offer better treatment to patients.
For instance, patients often forget their test results at home, or old tests get lost eventually. Imagine that all your medical records are stored in digital format, and when you go to Hospital A, a doctor there can examine your tomogram taken 4 years ago at Hospital B where your name was misspelled by the clerk
Future Development Goals
There are several improvements possible in the module. Some of these are:
UI improvements and bug fixes. Feedback from implementors and open tickets contain suggestions on how to improve web pages and work flow.
Metadata storage from matching runs. Currently, only patient matching output is stored. Information such as the user that ran the report, the runtime of the report, and which strategies were used should be associated for the output.
Integration into OpenMRS. The patient matching module might be moved from a module to a part of core OpenMRS. Code and resources will need to be organized, moved, and improved.
Downloads
http://modules.openmrs.org/modules/view.jsp?module=patientmatching
http://source.openmrs.org/browse/Modules/patientmatching/
https://github.com/openmrs/openmrs-module-patientmatching
What the module does
The OpenMRS module wraps around the patientmatching jar to facilitate creating the matching file setup and merging patients that are found to be duplicates
Terms and Concepts
The matching method used in the module is the Felligi-Sunter probabilistic matching. By knowing the agreement rates of specific fields between true and false matches, the likelihood of a given pair being a true match can be calculated, and a score is given based on how well the fields match.
The m-value is the rate a field is equal between true matches. As an example, sex might have an m-value of 0.90, meaning 90 percent of the time the value for "sex" will be the same if the two records are for the same person.
The u-value is the rate a field is equal between non-matches. As an example, sex might have a u-value of 0.5, meaning between two records' "sex" value will match about half the time. This is reasonable, as there are usually 2 values for "sex" and since the 2 values are close to equally prevalent, there is about a 0.5 chance that two different people will agree on that field. Similarly, month of birth might be close to 1/12th.
Blocking is the way to decrease the search space. It is possible to compare every record against every other, but the majority of the comparisons will be between records that are not matches, and the search space grows exponentially. A good set of blocking schemes should try to cover most of the true matches while minimizing extra work. In this program, blocking is done by choosing some fields to match and then comparing only pairs that have the same value for that field. Choosing last name as a blocking field, for example, would compare records that had the same last name but might differ in first name or medical record number. It would not, however, find the records where the last name was misspelled. Having a second blocking scheme that blocks on month of birth and date of birth might find this match if the errors do not include month and date of birth.
Difference between "Must Match" and "Should Match"
"Must Match" is used to determine which patients get compared to other patients. For the demographics selected, these values must match to have the module evaluate the records.
"Should Match" indicates the fields that contribute to the score. If the two patients are the same person, then these fields should match. The module will compare these values between patients, and based on the analysis, set the score.
What the standalone jar does
There is a main() method in the org.regenstrief.linkage.gui.RecMatch. The Java Swing GUI allows more options than the OpenMRS module, but does not work with data in an OpenMRS system. It matches records in flat database tables or delimited files.
Sample matching run using the GUI
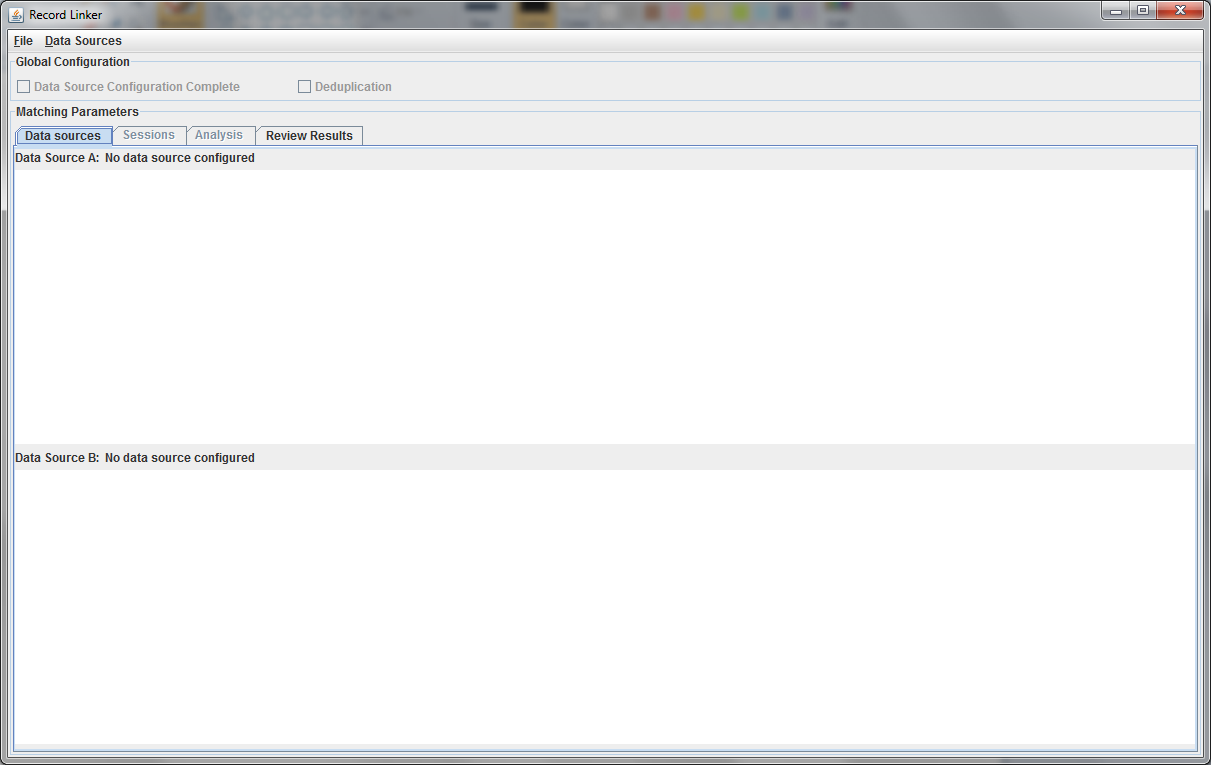
Upon startup, the application will look like below:
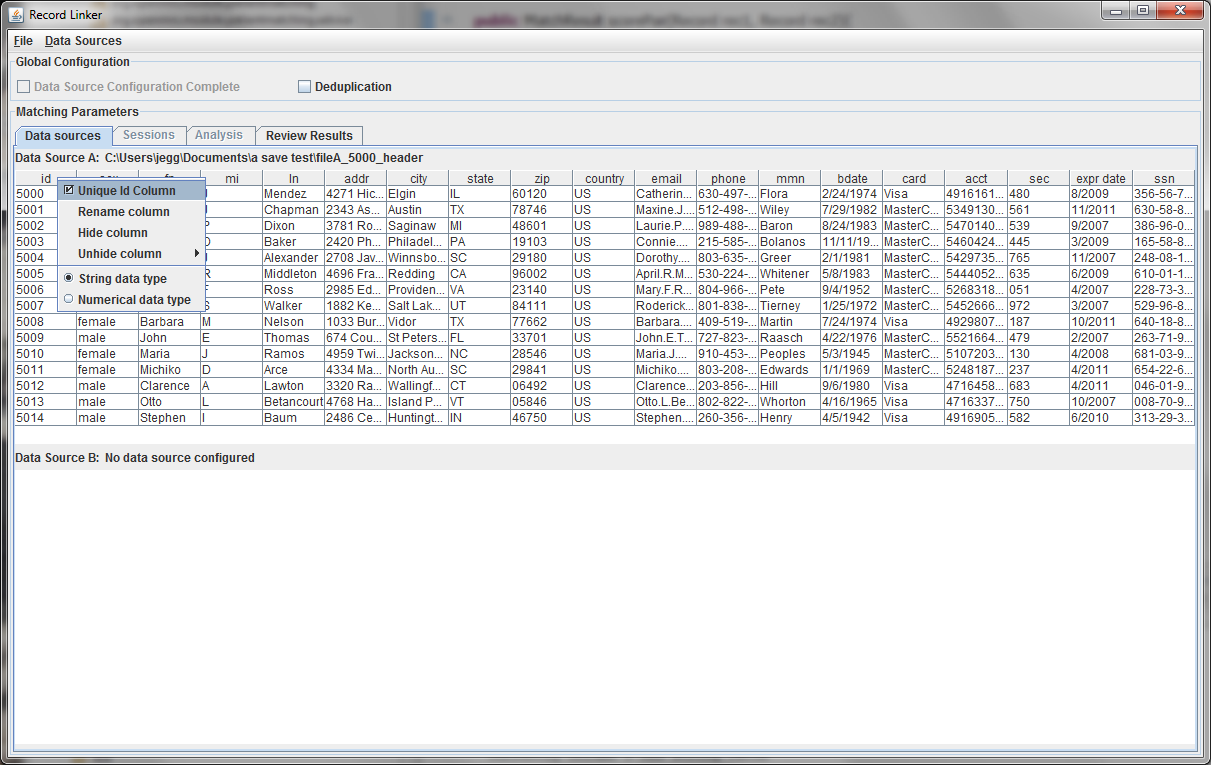
This initial tab is where the user specifies the data that will be matched. The "Data Sources" menu has options to configure the top and bottom. The top, datasource A, must be configured.
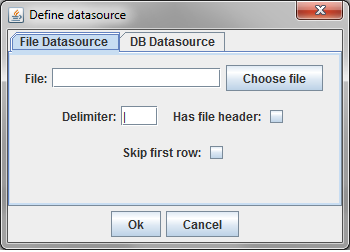
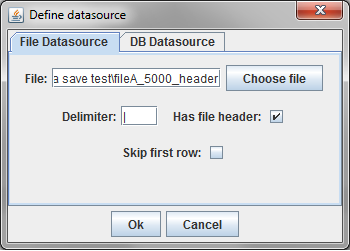
In this example, a delimited text file was chosen. The parsing of text files is very simple, and does not understand quoting strings. It is possible that preprocessing the data might be required.
The sample file chosen has a header row and uses the default '|' character to delimit fields. Since there is a header row, the demographic fields will be labeled as they are in the file. If there were no header line, the fields would be labelled starting with "Column A."
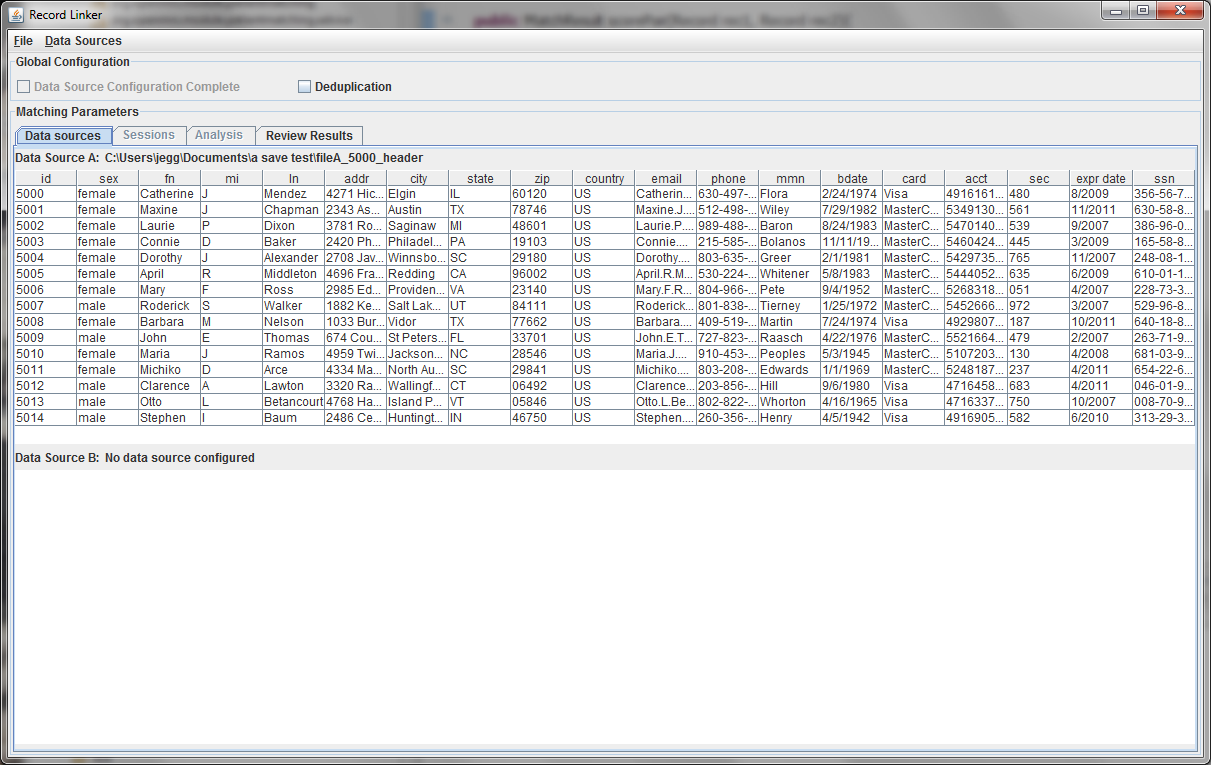
The first few lines of the file are displayed in a table. Right clicking on the table headers allow the user to change the names of the fields or specify that a numeric test of equivalence be used instead of a string test.
Every dataset must have a unique ID field that is an integer. Having such a field is very useful when when running analysis and working on matching results, so the program expects a suitable column. A field that is a line number can be added to the file to fulfill the requirement.
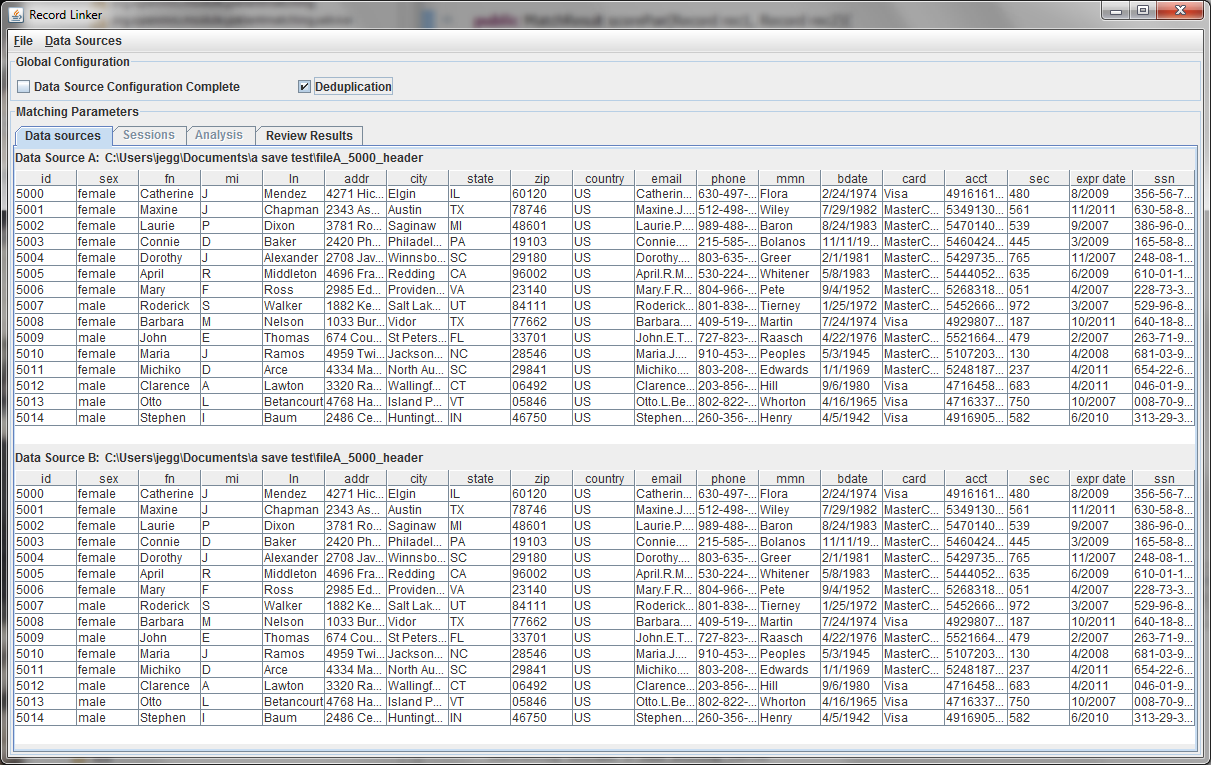
A second file can be specified using the "Data Sources" menu, or the check box for "Deduplication" can be selected. This will populate the bottom part with a copy of the top data, and duplicate records within the file will be found. Selecting "Data Source Configuration Complete" will move the user to the "Sessions" tab where matching options are set.
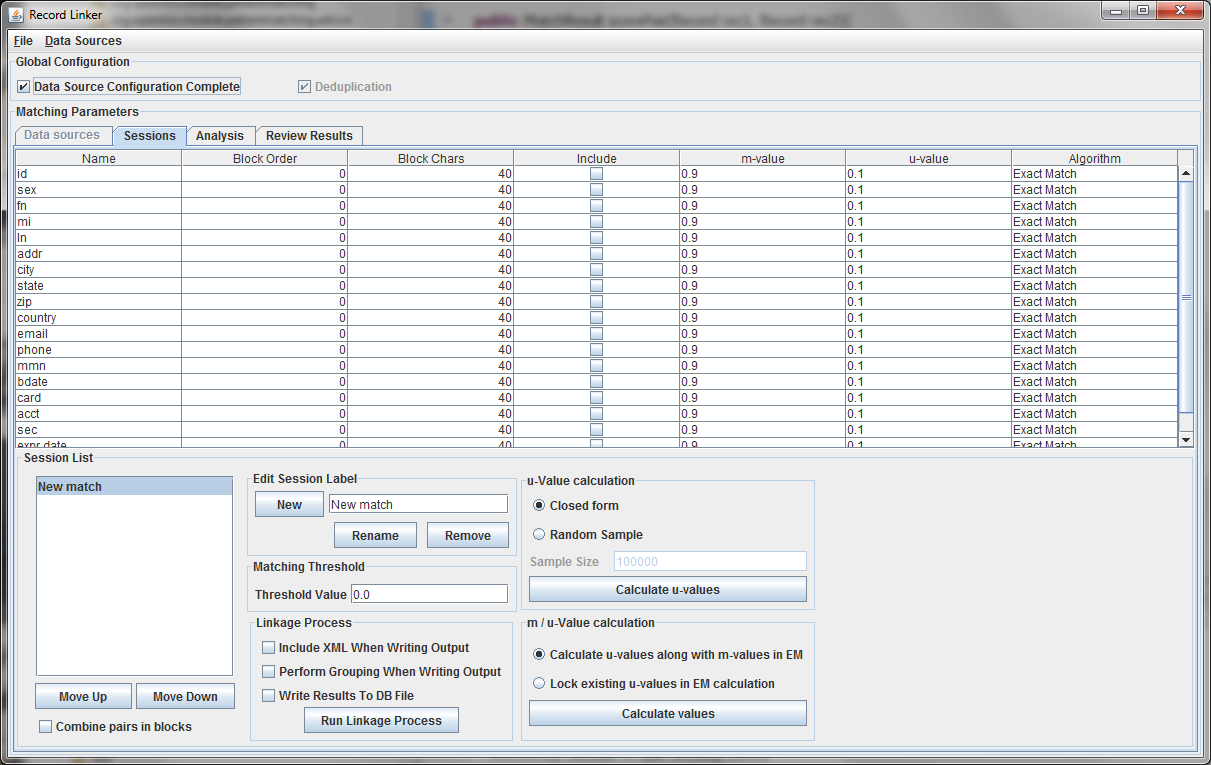
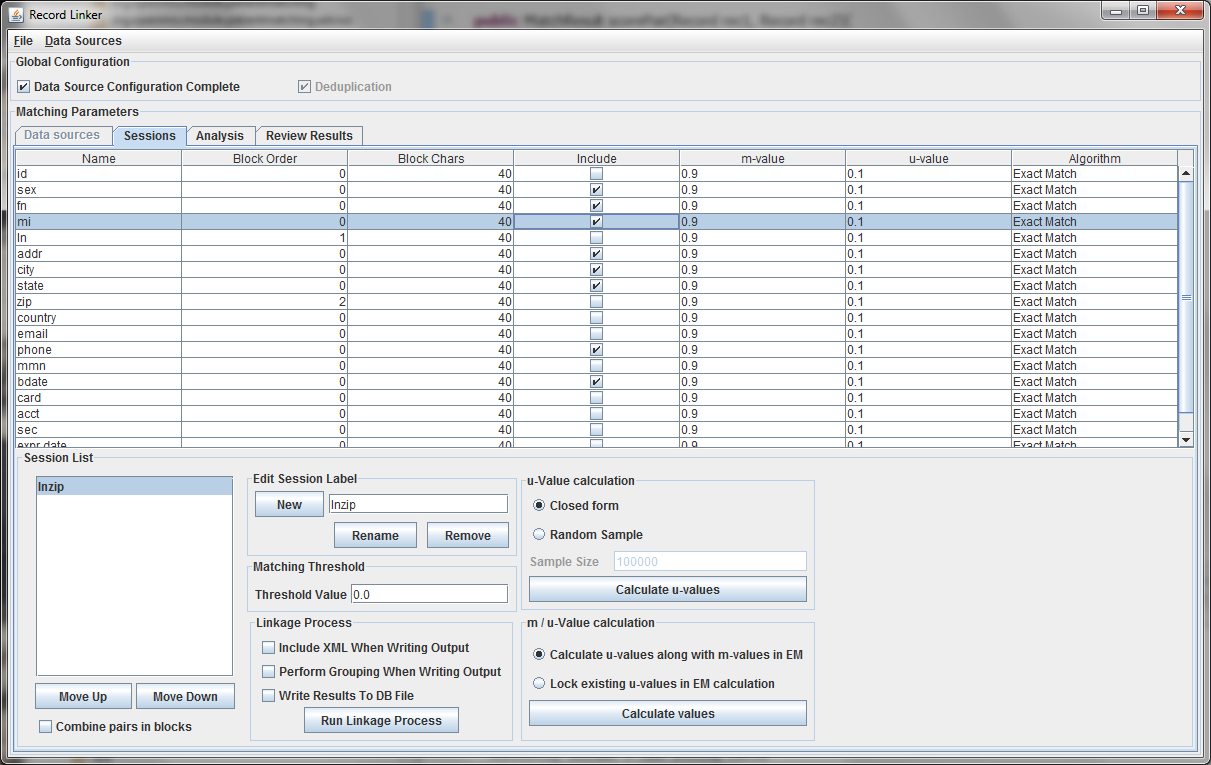
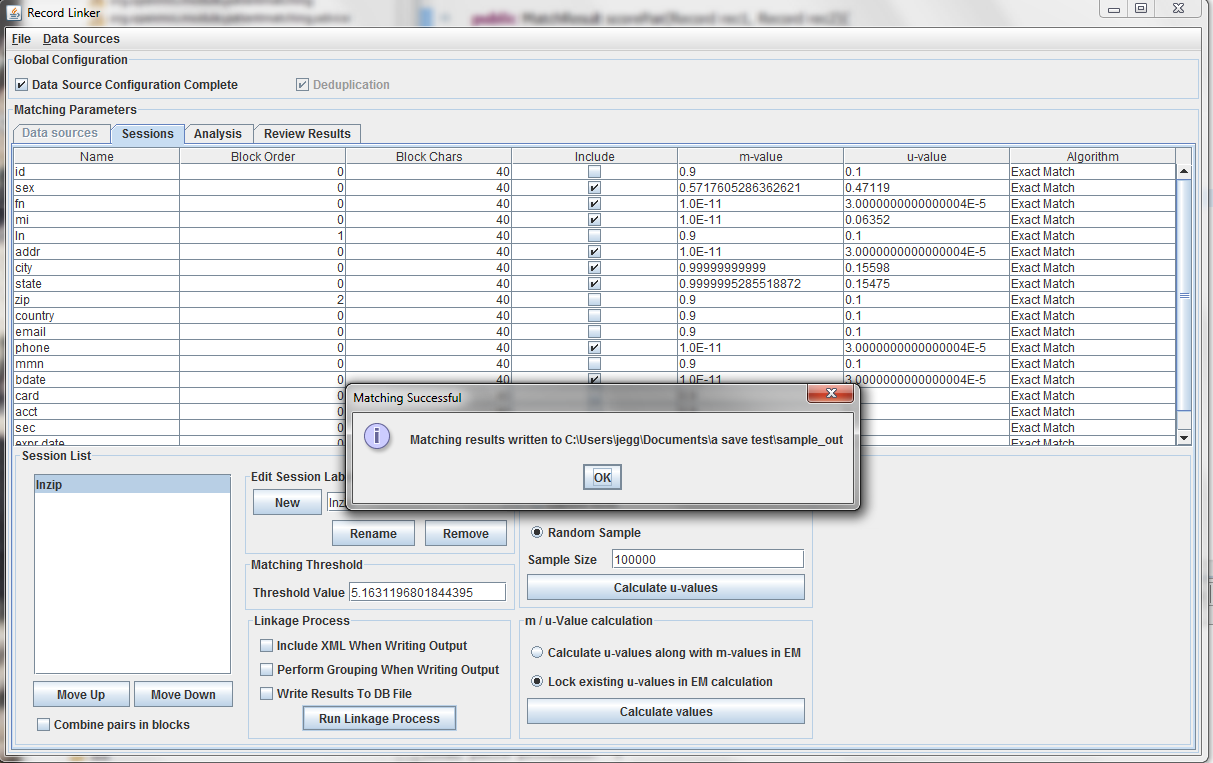
The table above shows the fields defined in the previous tab. The options and values are given for each field. The table displays the selected run defined in the box at the lower left. Multiple runs can be specified, since blocking on different columns will form different candidate pairs, and a few well chosen runs are better than a single inefficient run.
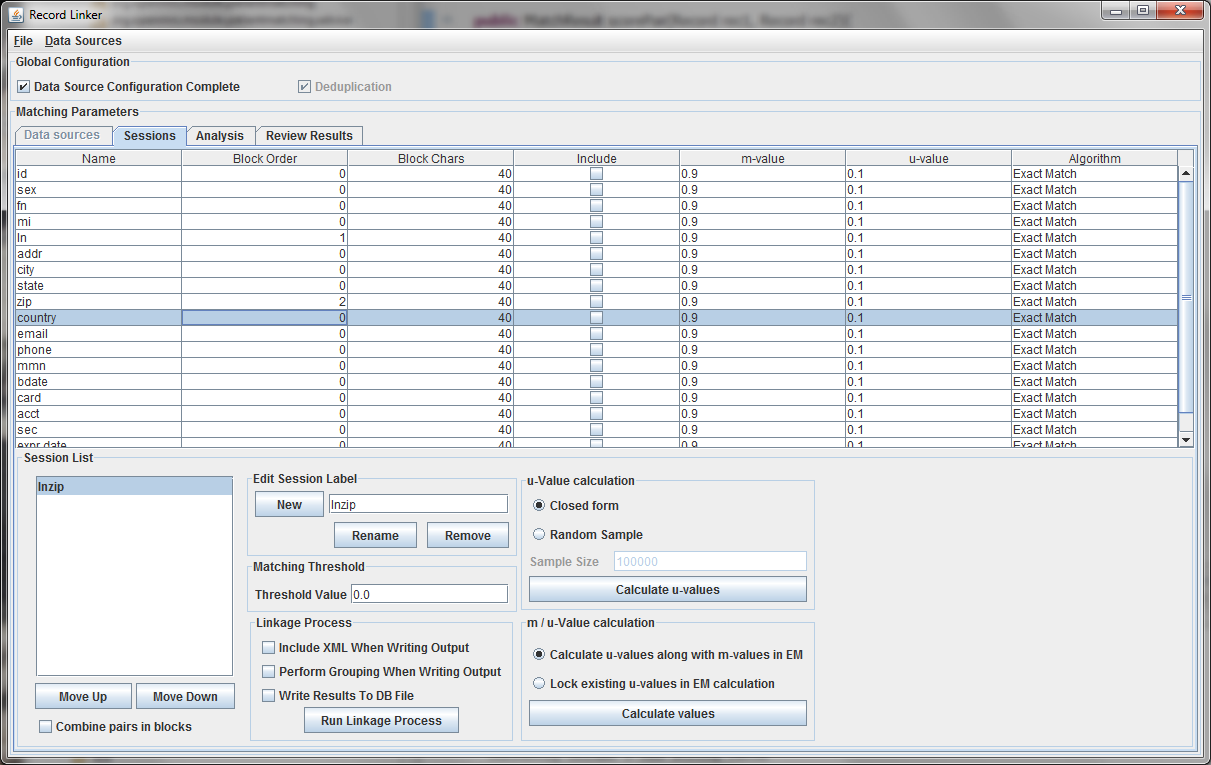
The session displayed has been renamed by typing "lnzip" in the text field and clicking "Rename." It is useful to choose descriptive names, and our convention has been to use the blocking columns as the name for a session, since blocking columns are the most important choice in a matching scheme.
The blocking columns, "ln" and "zip" have been set. Blocking columns are numbered sequentially starting from 1. The order does not matter, but the different values are used by the program to sort the data in order to run efficiently. The "Include" column shows which columns to use when making the comparison. For these columns, whether they match between pairs will increase or decrease the score. The blocking fields and the fields that are no selected for matching will be carried through to the output, but they will not affect the final score.
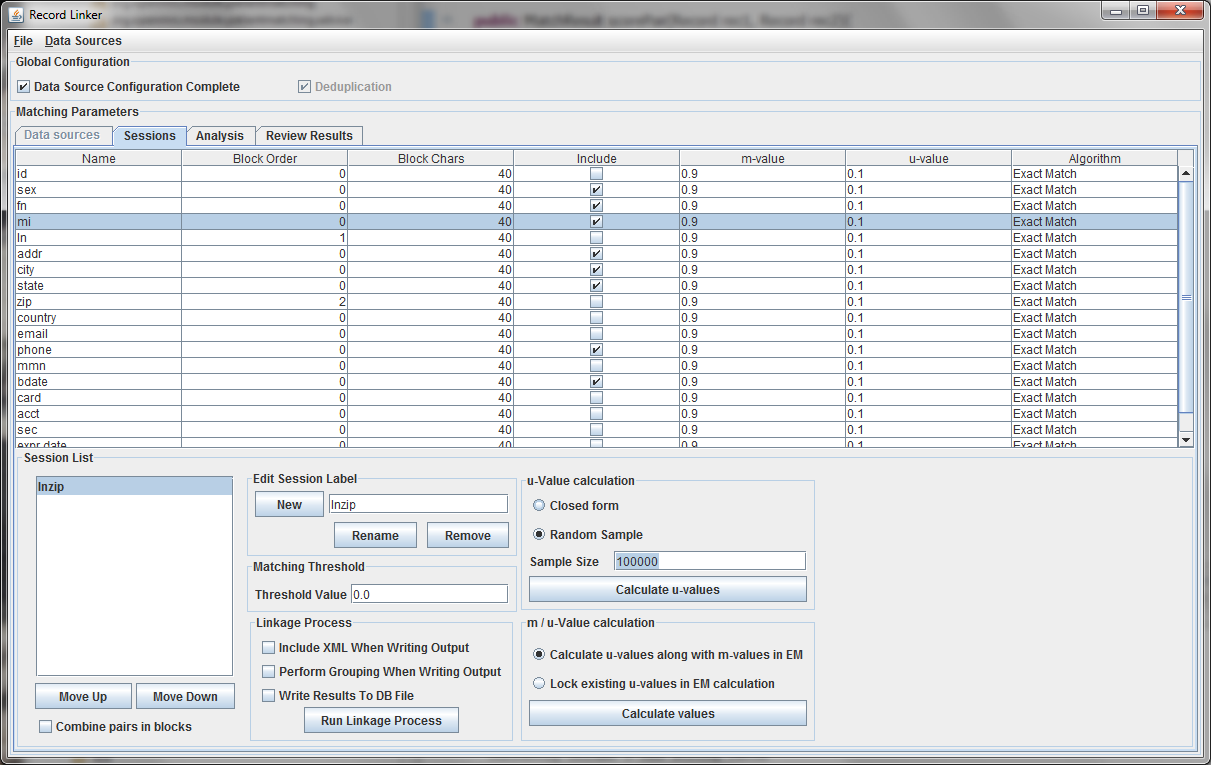
It is possible to calculate matches and write output in this state, but the default values of 0.9 and 0.1 are very likely inaccurate, and matching will be poor. Calculating the m-values and u-values will increase the quality of matching greatly, and random sampling is currently the standard way the program calculates u-values.
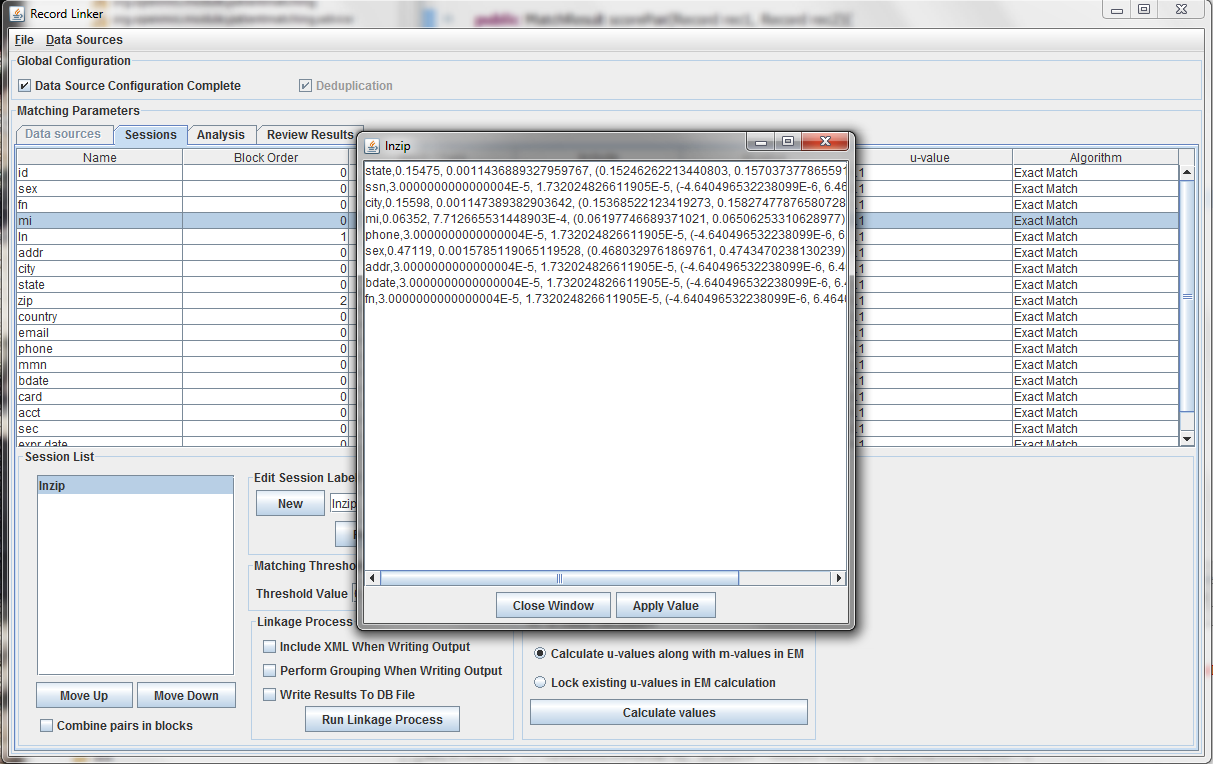
After selecting the "Random Sample" button and clicking "Calculate u-values" the analysis will be performed. The output will be shown, and clicking "Apply value" will modify set the run's u-values to what was calculated.
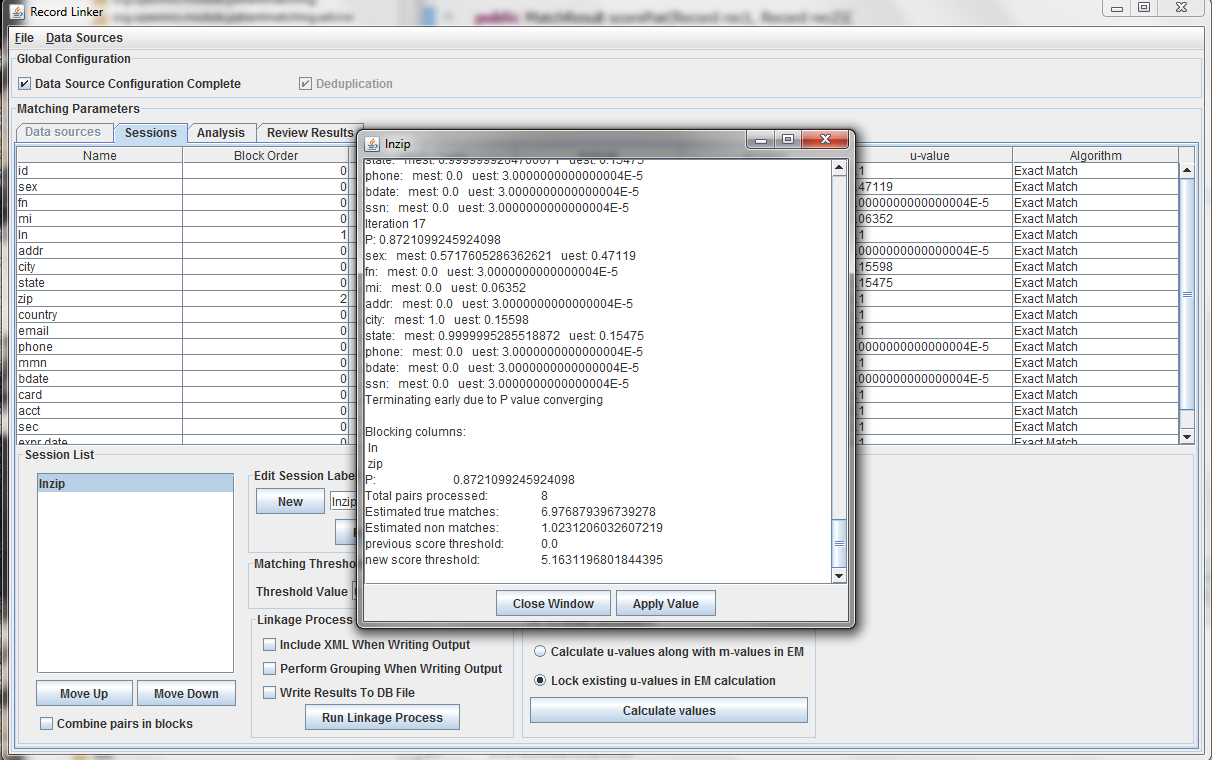
Expectation maximization (EM) provides a way to calculate either the m and u-values, or the u-values can be locked and EM will only calculate the m-values. Random sampling is a more accurate way of calculating u-values, so the current best method is using random sampling to calculate u-values and selecting "Lock existing u-values in EM calculation."
Clicking "Calculate values" will run the analysis and display the output. Clicking "Apply value" will set the run's values to what was calculated. The matching threshold is also calculated by EM, and that is also copied when "Apply Value" is chosen.
Clicking "Run Linkage Process" will create an output file with the pairs created by the blocking fields and the scores based on the m and u-values set for the include fields.
GUI Options
The number of "Block Chars" indicate how many initial characters to use when comparing for equivalence between blocking fields. If first name is a blocking field, then normally "Jenny" and "Jennifer" would not be equal and two records with those values would not be compared. If "Block Chars" were set at 4, then the first four characters, "Jenn," would be compared and the two records compared. This feature is not commonly used or well tested, and might have problems finding all the correct pairs.
The "Algorithm" option determines what test of equivalence to use when comparing fields. Blocking will always use "Exact Match" but fields included in scoring can use others. The options is a Levenshtein, longest common substring, or Jaro-Winkler similarity metric. The thresholds for the algorithms are 0.7, 0.8, and 0.8 respectively. Using these alternate comparisons might compensate for misspellings in the data.
The option "Combine pairs in blocks" passes through all unique pairs of all blocking runs that are defined and treats them the same. Instead of calculating m-values and u-values separately, this will calculate aggregate statistics for all runs. It will use all pairs when writing output, and use a common scoring scale and threshold. Currently, only the selected run is changed to reflect the analysis results.
"Include XML When Writing Output" will write an XML file along with the standard text file. This XML file has more information such as which fields were considered matches, what algorithm was used, what contribution to the score each field gave, etc.
The option "Write Results To DB File" will write the matching output to an SQLite database file that can be opened in the "Review Results" tab.
The "Closed Form" method to calculate u-values calculates the correct u-values directly using the frequency of values in the data. This method uses much more memory and could take longer than random sampling.